Photo by Thought Catalog on Unsplash
We are seeing a lot of contents from Massive Open Online Courses (MOOCs) and open sourced communities by researchers or academics in the field. That, and most of the contents are available at no cost, makes it an attractive way for students, academia, or even just regular people who want to increase their knowledge in the field to learn and polish their skills in the field.
The contents and materials aren’t even bad like one might think for free content. In fact, a lot of the courses from MOOCs platforms come from partnering with major universities such as Harvard (edX) and Johns Hopkins (Coursera). However, there are some downsides to joining these free classes, though. Most of the time, you can’t access certain course materials, join assignments or quizzes, and receive certificates. Still, it’s worth taking to help you get a good start in the field you’re looking to improve. That way, you can explore and put your newfound skill into practice and build your own portfolio from the ground up, which equally attracts talent searchers.
By the way, some of the resources here include links to Youtube playlists from lecturers of representative universities. And since these times call for mostly online based learning, you can use this tool Prodeus to keep track of your learning progress and get your degree from Youtube (Yes, that’s even their tagline).
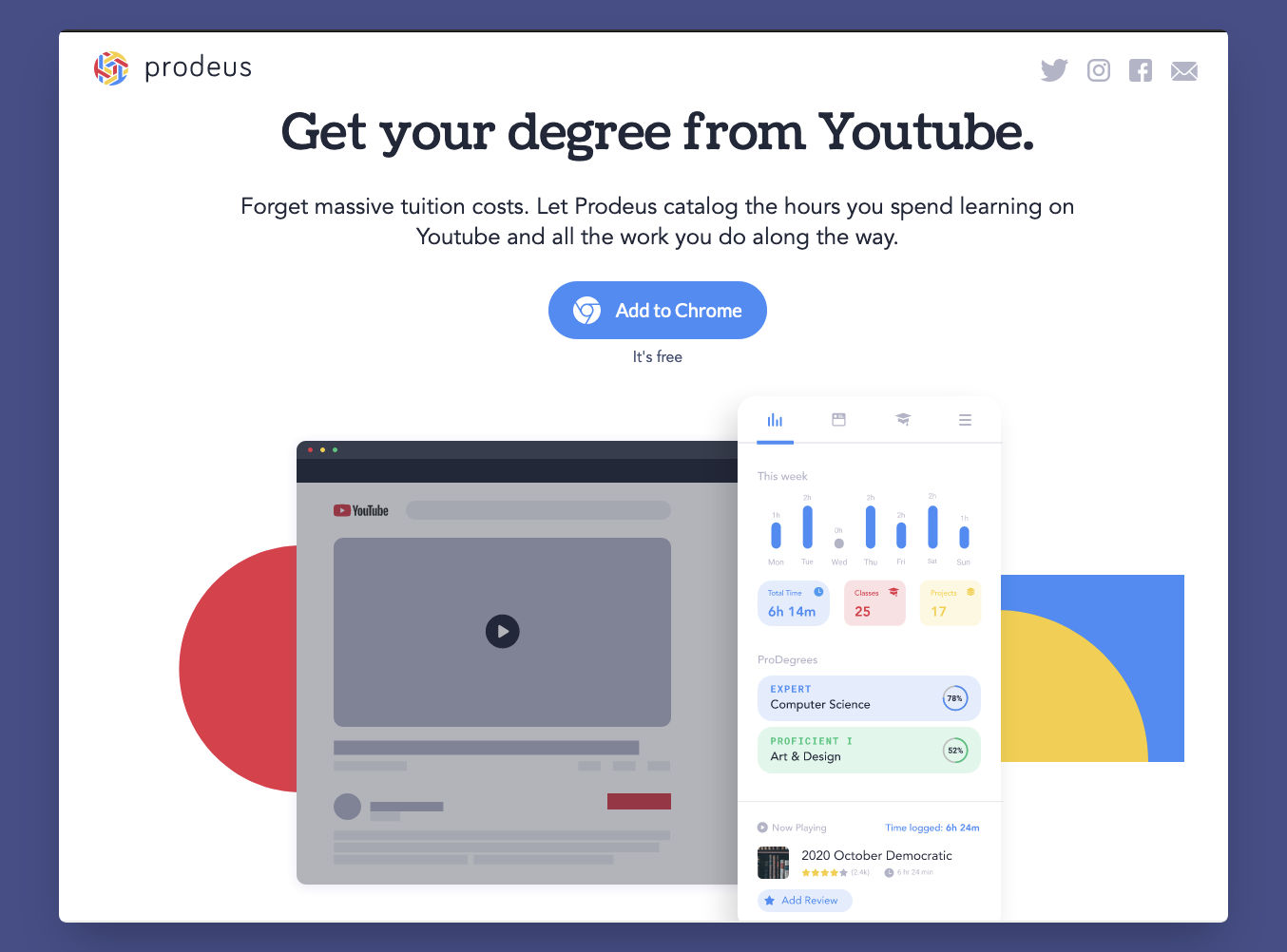 Prodeus landing page
Prodeus landing page
Hope this helps! We’ll cover some other productivity tools to help your learning journey in the next articles.
Notable Sources
There is actually a lot of resources out there that you can use to learn Bioinformatics either for free or paid. One of the most notable one comes from two sources listed below.
OSSU for BioInformatics
The Open Source Society Curriculum for Bioinformatics
This is a open sourced, community driven curriculum with online resource collections to learn Bioinformatics for free. Their description is :
This is a solid path for those of you who want to complete a Bioinformatics course on your own time, for free, with courses from the best universities in the World.
The length of the course here is meant to be taken for in 4 years. But there is a case of a guy that went through learning an entire 4 year MIT curriculum for Computer Science in 1 year, so I guess it depends on how motivated or ambitious you are as a person.
Here is an excerpt from the curriculum from OSSU for Bioinformatics.
Sample Curriculum for 2nd Year 🔖
| Code | Course | Duration | Effort |
|---|---|---|---|
| BIO 2311 | Biochemistry | 15 Weeks | 4-6 Hours/Week |
| CHEM 2311 | Organic Chemistry | 15 Weeks | 4-6 Hours/Week |
| COMP 2311 | CS 2 - Object Oriented Java | 6 Weeks | 4-6 Hours/Week |
| MATH 2311 | Calculus 3 - Integration | 4 Weeks | 8 Hours/Week |
| MATH 2312 | Mathematics for CS | 13 Weeks | 6 Hours/Week |
| COMP 2312 | Introduction to Databases | 10 Weeks | 8-12 Hours/Week |
| MATH 2313 | Linear Algebra | 15 Weeks | 8 Hours/Week |
| COMP 2313 | Introduction to Linux | 8 Weeks | 5-7 Hours/Week |
| MATH 2314 | Inferential Statistics (with R) | 5 Weeks | 6 Hours/Week |
📖 I suggest you go ahead and head over to their GitHub page to learn more about the curriculum.
Rosalind.info
 Rosalind info logo
Rosalind info logo
Here’s something a bit more interesting. For those familiar with programming tutorials and hands-on exercises on the web, you’re probably familiar with learning projects such as Project Euler. Students or other users can test their ability to solve a series of challenging problems that test their practical understanding rather than theoretical knowledge. The Rosalind Project is similar to Project Euler, but in this case, it is more focused on the Bioinformatics side.
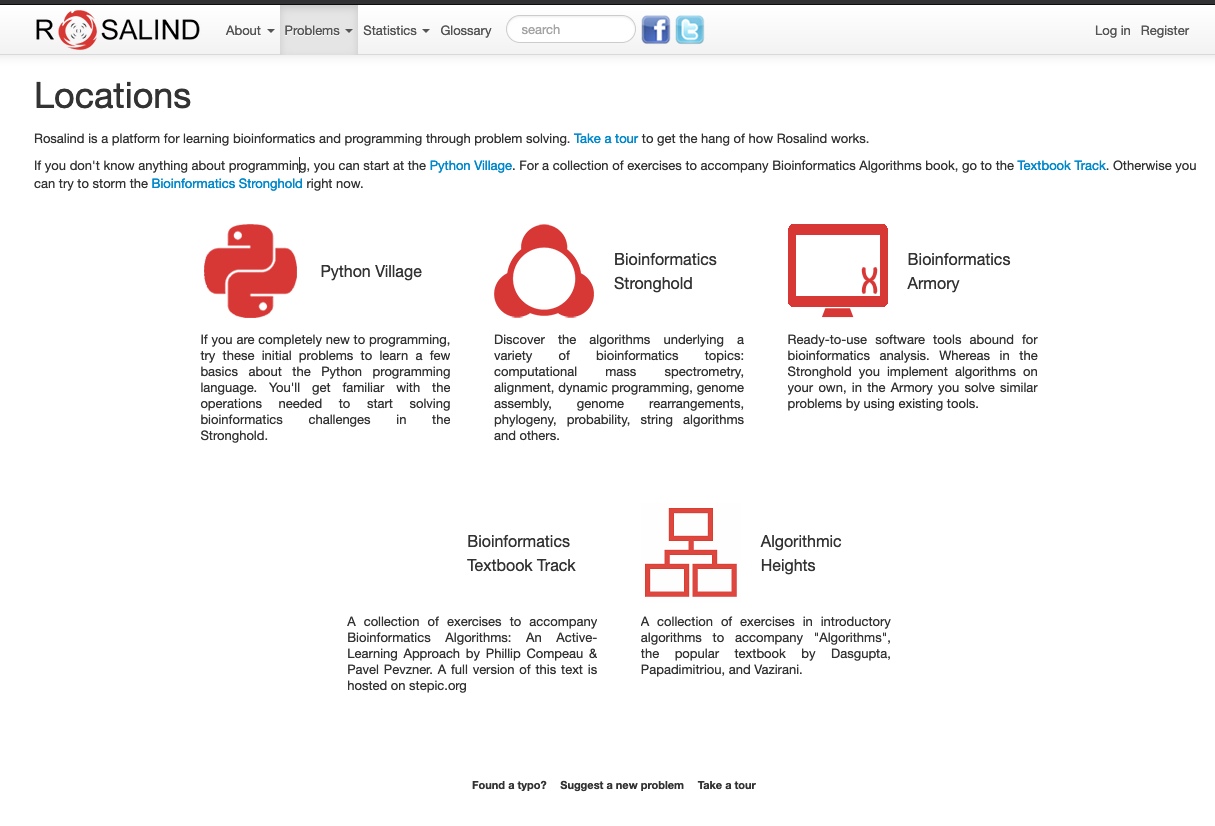 Rosalind landing page
Rosalind landing page
Here you can test your programming knowledge and skills along with several other supporting materials for Bioinformatics, such as:
- Tools used in Bioinformatics
- Textbooks
- And Introductory lessons to algorithms such as graph theories and many more
Other Sources
The two previously mentioned resources here are useful to get you a clear general view on Bioinformatics and acts as guidelines for you to start learning on your own both in terms of theoretical and practical knowledge.
However, if, for some reason, those are not cut out for you, there are many more resources out there that you can use as well. I’ll list it down below based on the category. Also, since this article focuses more on Bioinformatics resources, other resources for other topics will be written separately in the next article. Enjoy ☀️!
Biology 🌏
🎒 Here you’ll find a list of online learning courses for Bioinformatics.
| Course | What it’s about |
|---|---|
| Essential Human Biology: Cells and Tissues | An edX course on the introduction and foundation of the human cells and tissue structures and functions |
| Tales from the Genome | This is a course from Udacity by the 23andMe company focusing on introductory lessons to Genetics for beginners |
| Bioinformatics Specialization | A coursera specialization track for those who are interested to learn deeper about Bioinformatics |
| Genomics Data Science Specialization | A coursera specialization track for you to understand more about genomics data from next generation sequencing (NGS) experiments. This specialization uses hands on exercises with Python, R, Bioconductor, and Galaxy. (I’ll leave more about this in the next article as well) |
| Systems Biology and Biotechnology Specialization | A coursera specialization where you will learn methodologies in Bioinformatics, Dynamic Modeling, Genomics, Network and Statistical Modeling, Proteomics, and Omics technologies |
| Bioinformatics Algorithm | This is an interactive content from the “Bioinformatics Algorithm” textbook co-written by the creator of Rosalind |
| Genomics Medicine Gets Personal | An edX course for introductory lessons to genomic medicine and personal genomic information |
📝 Other than the mentioned courses, there are also articles or Web Pages that can help you get a broader understanding of the subjects, along with some tutorials to help you get started.
| Resource | Contents |
|---|---|
| One Stop Data Analysis | This is a Web page with a compilation of tutorials for Bioinformatics analysis and many more |
| Biology for Computer Scientist | This is a GitHub repo to briefly learn about biology, immunology, along with the protocols and tools used such as Igv (Integrated Genome Browser) |
| The Genetic Code | This is a brief article describing DNA which I found to be quite informative |
| Introduction to Applied Bioinformatics | This is a free interactive article that introduces key concepts Bioinformatics in terms of implementation and applications |
| Microbial BioInformatics Starter Kit | This article consists of list of tutorials and help pages for Bioinformatics I think you’ll find useful. This is written by Hanage Lab the Harvard TH Chan School of Public Health |
| Computational Genomics Class | This is a compilation of video lessons from Dr. Rob Edwards from San Diego State University. The contents are bite sized information on NGS technologies, Bioinformatics algorithms |
🏫 There are also several universities that post their lectures online for the public. You can find the lectures in document format though some are available as video lectures as well.
| University | Lecture types |
|---|---|
| MIT Opencourseware | This is a course in Fundamentals of Biology by the MIT. You can learn Biochemistry, Molecular Biology, Genetics, and Recombinant DNA |
| Cornell University Institute for Computational Biomedicine | This is a course for Essentials of Bioinformatics, however, most of the materials are links to relevant publications instead of lecture notes |
| University of Washington Computer Science and Engineering | The courses here includes lecture notes, some useful links, and also biological animations such as DNA models |
| NPTEL Courses in Bioinformatics Algorithm and Applications | This is a video lecture series in Bioinformatics algorithm that you can watch, download and gain access to lecture materials and assignments |
| Norwegian Veterinary Institute BioInformatics Training | This is an online GitHub repo for BioInformatics training |
Computation 💻
Before we get into the resources here, I would like to mention that when you want to work on biological data, it is easier for you to use the already established tools available. One of which is Bioconductor, one of the providers for open source software for Bioinformatics for R. You can even find a course for Bioconductor here from Harvard T.H. Chan School of Public Health.
For those who are more into Python, there is also a library of tools for computational molecular biology from Biopython. There are many more libraries out there that you can use, but these are the common ones. If you find one that you think is useful, you can go ahead and contribute to the repo.
🎒 Anyway, here’s some list of courses you can follow for computational biology and some programming. Most of them are interactive based tutorials that you can follow. I’ll do my best to categorize them based on their programming languages, starting from Python :
| Python Resources | What it’s about |
|---|---|
| Programming Course Syllabus | This is a compilation of resources to learn programming in Python that you can use on your own pace. Do keep in mind that some of the resources here are paid |
| Introduction to Interactive Programming in Python | This is a coursera course for those with little to no programming background to learn the basic of building simple interactive python application |
| Google’s Python Class | This is a free class for those who have little to no programming experience to learn Python. From google of course |
🖱️ Here are some courses for those wanting to learn R :
| R Resources | What it’s about | ||
|---|---|---|---|
| Swirl | “Learn R, in R”. Get yourself familiar with the inner workings of R… in R. | ||
| Introduction to R | This is a compilation of resources to learn R based on lecturer slides and notes from the King’s College of London | ||
| Codecademy R Course | This is a Codecademy course for R. Like Coursera and edX, you can enroll for free, but the content is limited and you won’t get a completion certificate | ||
| Data Mentor - Learn R Programming | This is a complete guide to learn R programming that includes the tutorials for installing the perquisites to run R in your local computer | ||
| R by Example | This is a collection of use cases of R based on real examples. Each examples contain source codes that you can copy and paste for you to try it out yourself in your local computer | ||
| R Markdown | This is a tutorial that focuses on helping you to create a reproducible documentation of your R code |
👨💻 Here are some courses for those trying to use the command line
| Unix / Command Line Resources | What it’s about |
|---|---|
| Linux Command Line Basics | Here are some collection to understand the basics of Linux command lines starting from setting up your Linux environment |
| Beginner’s Guide to Bash Terminal | This is a Youtube video to learn more about Bash terminal for Linux beginners |
| 8 Useful Shell Commands for Data Science | Here’s an article that sums up some of the common commands used in shell for Data Science |
| The Command Line | In here, you’ll find some introductory lessons to the command line along with some of it’s use cases in Bioinformatics, such as genome annotations, RNA-Seq, and many more |
| Linux Command Line Exercises for NGS data Processing | This contains tutorials and protocols using the command line for dealing with NGS data such as reading FASTA files, reading alignments score for Metagenomics / Population Genetics, and many more |
Mathematics and Statistics 📊
| Course | What it’s about |
|---|---|
| Mathematical Biostatistics Boot Camp 1 | This is a Coursera class focusing on the fundamentals of probability and statistical concepts in beginner data analysis |
| Models, Inference and Algorithms | This is a youtube playlist managed by the Broad Institute as an initiative to support the learning of biology, mathematics, statistics, machine learning, and computer science |
BioInformatics Tools and Supporting Materials
🎒 This is another resource list of supporting materials outside of courses. This includes tools and software, curriculum, or learning guidelines.
| Tools / Supporting Material | What it’s about |
|---|---|
| Bioinformatics Career Guide | Here you can find a more comprehensive list of courses for BioInformatics, however, note that some of the courses listed here are not free |
| Online BioInformatics Curriculum | This is a compilation of BioInformatics Curriculum from PLOS Computational Biology Journal |
| Computational Biology Curriculum | This is another compilation of Curriculum for Computational Biology published in PLOS Computational Biology |
| Guide to BioInformatics Tools | This article contains a list of commonly used BioInformatics tools |
| DNA Subway | This is a GUI based BioInformatics tool for Gene Annotations and Genome Analysis. Good for beginners who aren’t familiar with programming to get to know the workflow in BioInformatics analysis |
| Biocontainers | A containers that helps you streamline Bioinformatics workflow and packages. This is a bit advanced and requires some knowledge in container apps such as _Docker_, but is very useful to help your workflow by getting pre existing ones by the community |
| Explain Shell | This is a useful tool to help you guys understand the commands in command line in a visual way |
| Zero2bioinfo | This is a GitHub repo filled with tutorials to teach beginners some basic R and common Bioinformatics workflow. It also have some interactive exercises that you can play around with |
Closing
I know these resources aren’t as complete. Hopefully, more resources can be added to the next articles. Other than that, if you’d like to contribute, fork this GitHub repo and help expand the resources and make it a better place 🌟!
